Introduction
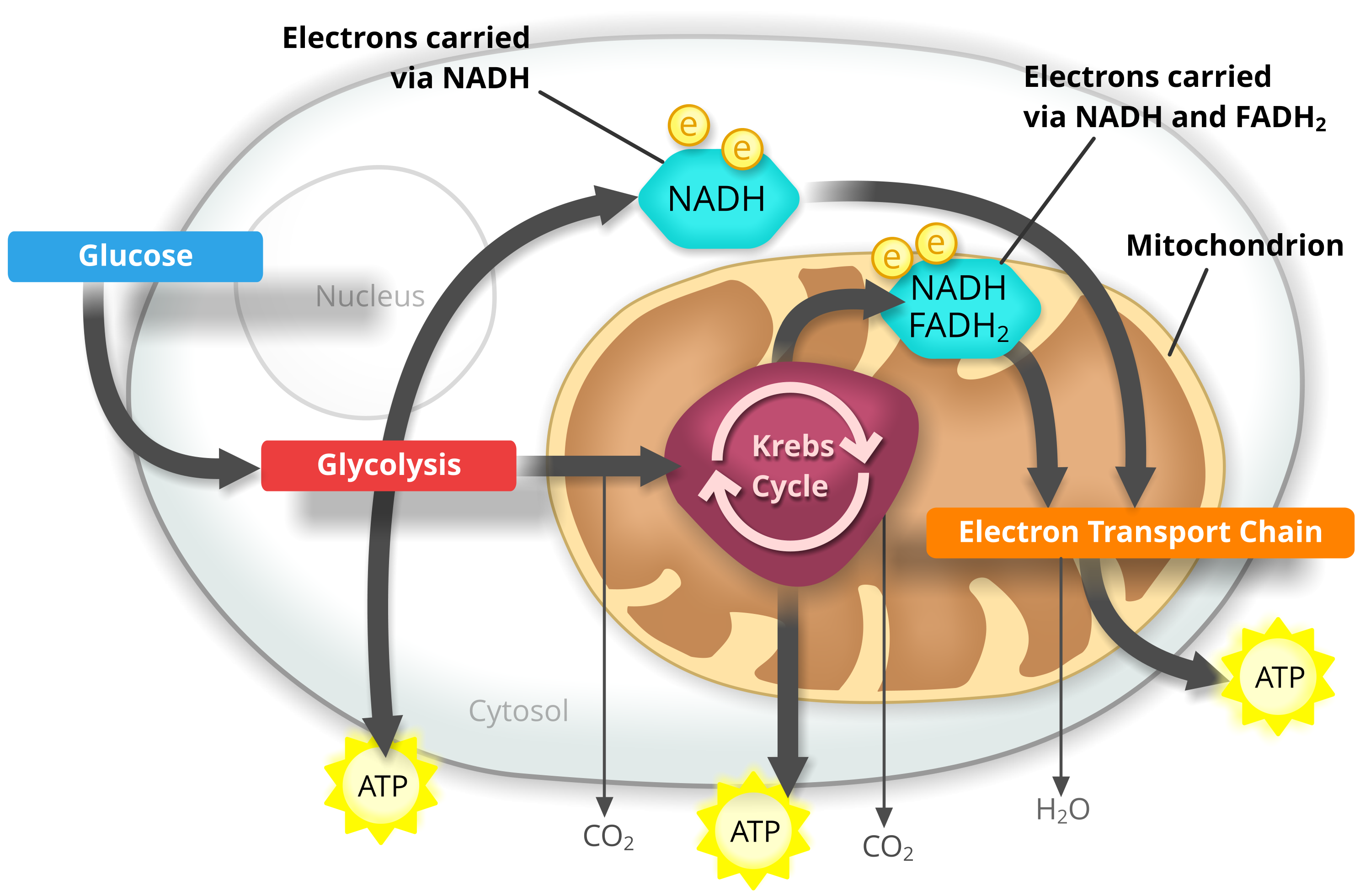
During cellular respiration in general and electron transport chain in particular, flavin adenine dinucleotide (FAD) plays a vital role. Figure 1 shows the workflow of cellular respiration in which the electrons (using nicotinamide adenine dinucleotide (NAD) and FAD) transfer from glucose (in glycolysis) to Krebs cycle, and the electron transport chain. Initially, the glucose undergoes a chemical transformation and is converted to pyruvate, a three-carbon organic molecule. During this glycolysis, ATP is generated, and NAD collects two electrons and two H+ molecules to transform to NADH. Each pyruvate from the mitochondrial process enters the mitochondrial matrix - the innermost compartment of mitochondria. In there, it is converted to a two-carbon molecule that binds to Coenzyme A, known as acetyl CoA, and releases carbon dioxide and produces NADH. The next process occurs within the Krebs Cycle, which regenerates four-carbon molecules and produces ATP, NADH, and FADH2 while releasing carbon dioxide. NADH and FADH2 entrust their electrons to the still-electron transfer chain, causing them to return to the "empty" form (NAD+ and FAD) to be ready for a new cycle. Electrons travel down the chain, releasing energy, and it is used to pump protons out of the mitochondrial matrix. The protons flow back into the substrate through an enzyme called ATP synthase, creating ATP. At the end of the electron transport chain, oxygen accepts electrons and occupies protons to form water.
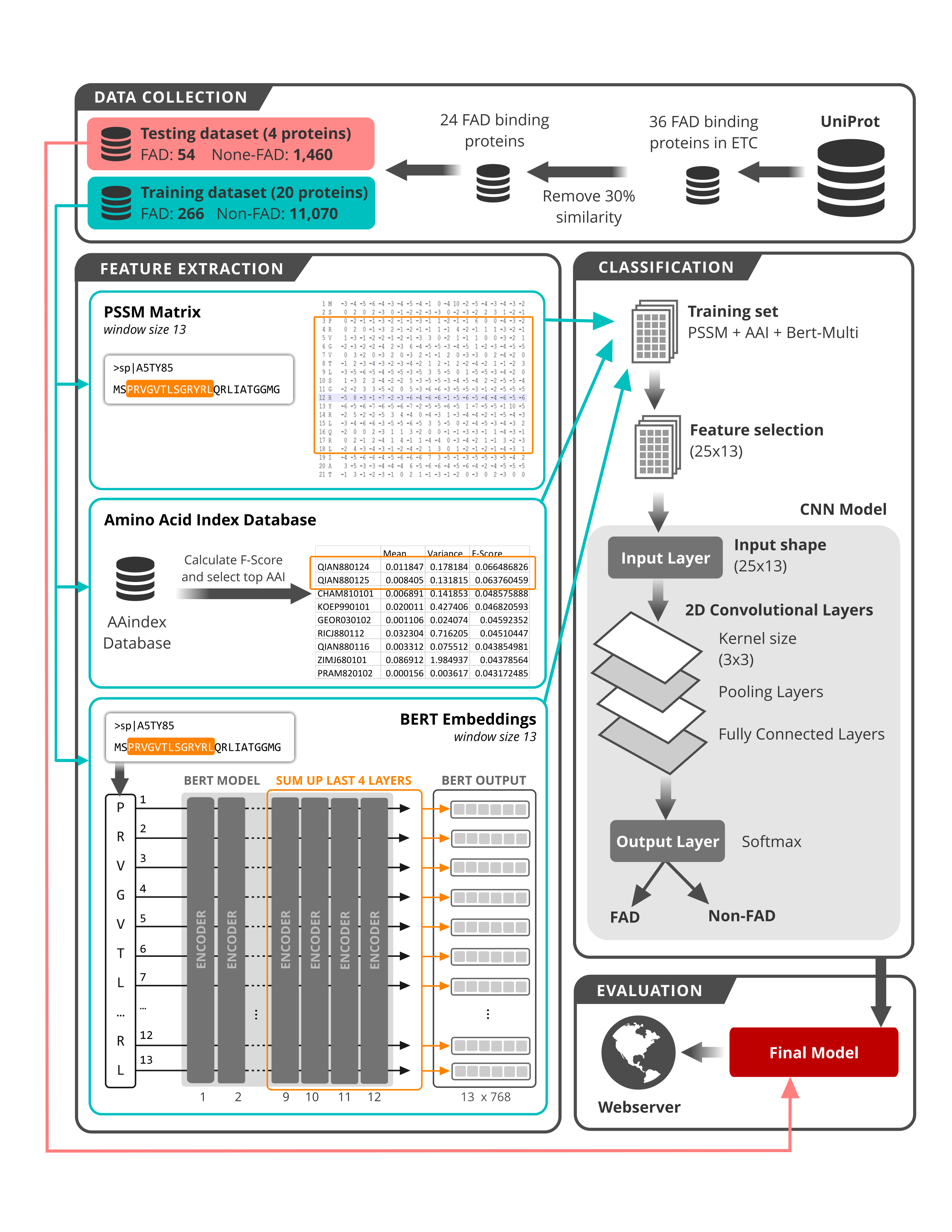
Electron transport chain is a series of protein complexes embedded in the process of cellular respiration, which is an important process to transfer electrons and other macromolecules throughout the cell. Identifying FAD binding sites in the electron transport chain is vital since it helps biological researchers precisely understand how electrons are produced and are transported in cells. This study proposes a new approach based on Pre-training of Deep Bidirectional Transformers (BERT), PSSM profiles, AAIndex to predict FAD binding sites from newly found naturally occurring electron transport proteins. Figure 2 shows the detailed workflow of this study.
Dataset
The dataset used in this server were retrieved from UniProt. The detail of the dataset is listed in the table below.
| Original | Similarity < 30% | Training | Independent | |
|---|---|---|---|---|
| FAD binding proteins | 36 | 24 | 20 | 4 |
| FAD binding sites | 523 | 320 | 266 | 54 |
| Non-FAD binding sites | 21,983 | 12,530 | 11,070 | 1,460 |
| Total | 22,506 | 12,850 | 11,336 | 1,514 |
If you would like to build a model and evaluate our model, we provide the dataset as the below link.
Download dataset.zipResults
Our proposed approach archived 85.14% accuracy and improved accuracy by 11%, with Matthew's correlation coefficient of 0.39 compared to the previous method on the same independent set. Furthermore, we distilled and examined contextualized word embedding from pre-trained BERT models to explore similarities in natural language and protein sequences
| TP | FP | TN | FN | Sens | Spec | Acc | MCC | |
|---|---|---|---|---|---|---|---|---|
| PSSM | 44 | 255 | 1205 | 10 | 81.48 | 82.53 | 82.50 | 0.30 |
| PSSM + AAI | 45 | 247 | 1213 | 9 | 83.33 | 83.08 | 83.09 | 0.31 |
| PSSM + BERT-Base | 44 | 253 | 1207 | 10 | 81.48 | 82.67 | 82.63 | 0.30 |
| PSSM + BERT-Large | 44 | 4243 | 1217 | 9 | 81.48 | 83.36 | 83.29 | 0.31 |
| PSSM + BERT-Multi | 45 | 239 | 1221 | 9 | 83.33 | 83.63 | 83.62 | 0.32 |
| PSSM + AAI + BERT-Large | 46 | 221 | 1239 | 8 | 85.19 | 84.86 | 84.87 | 0.34 |
| PSSM + AAI + BERT-Multi | 46 | 217 | 1243 | 8 | 85.19 | 85.14 | 85.14 | 0.34 |
Submission
In order to avoid the errors, please submit the sequence in fasta format (we also give you the fasta file examples). The user can choose two options to submit, including paste the sequence into text area and upload sequence file. The user can submit one single fasta file or multiple fasta file. In the result page, we show the results for the sequences which belong to electron transport proteins or not.
Sample fasta Sequence(s)>sp|P00455|FENR_SPIOL Ferredoxin--NADP reductase, chloroplastic OS=Spinacia oleracea OX=3562 GN=PETH PE=1 SV=1 MTTAVTAAVSFPSTKTTSLSARSSSVISPDKISYKKVPLYYRNVSATGKMGPIRAQIASD VEAPPPAPAKVEKHSKKMEEGITVNKFKPKTPYVGRCLLNTKITGDDAPGETWHMVFSHE GEIPYREGQSVGVIPDGEDKNGKPHKLRLYSIASSALGDFGDAKSVSLCVKRLIYTNDAG ETIKGVCSNFLCDLKPGAEVKLTGPVGKEMLMPKDPNATIIMLGTGTGIAPFRSFLWKMF FEKHDDYKFNGLAWLFLGVPTSSSLLYKEEFEKMKEKAPDNFRLDFAVSREQTNEKGEKM YIQTRMAQYAVELWEMLKKDNTYFYMCGLKGMEKGIDDIMVSLAAAEGIDWIEYKRQLKK AEQWNVEVY >sp|P00221|FER1_SPIOL Ferredoxin-1, chloroplastic OS=Spinacia oleracea OX=3562 GN=PETF PE=1 SV=2 MAATTTTMMGMATTFVPKPQAPPMMAALPSNTGRSLFGLKTGSRGGRMTMAAYKVTLVTP TGNVEFQCPDDVYILDAAEEEGIDLPYSCRAGSCSSCAGKLKTGSLNQDDQSFLDDDQID EGWVLTCAAYPVSDVTIETHKEEELTA
